Scalable Bioinformatics Services
I have previously [Bailey and Machanick 2012, Machanick and Bailey 2011, Bailey et al. 2010] worked on the
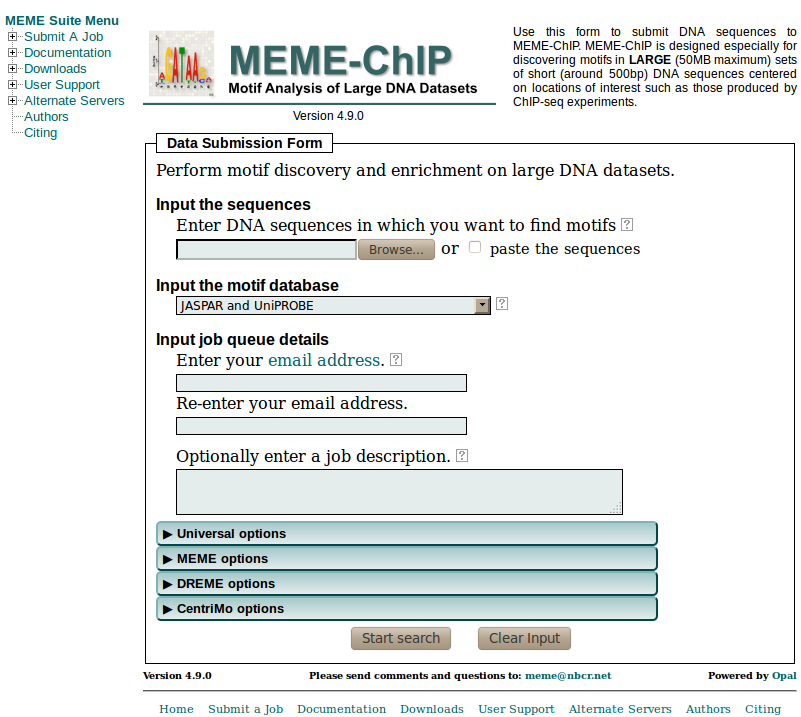
MEME suite of web services, which encompasses
a wide range of bioinformatics tools with an emphasis on DNA sequence analysis (though some tools also support proteins). The
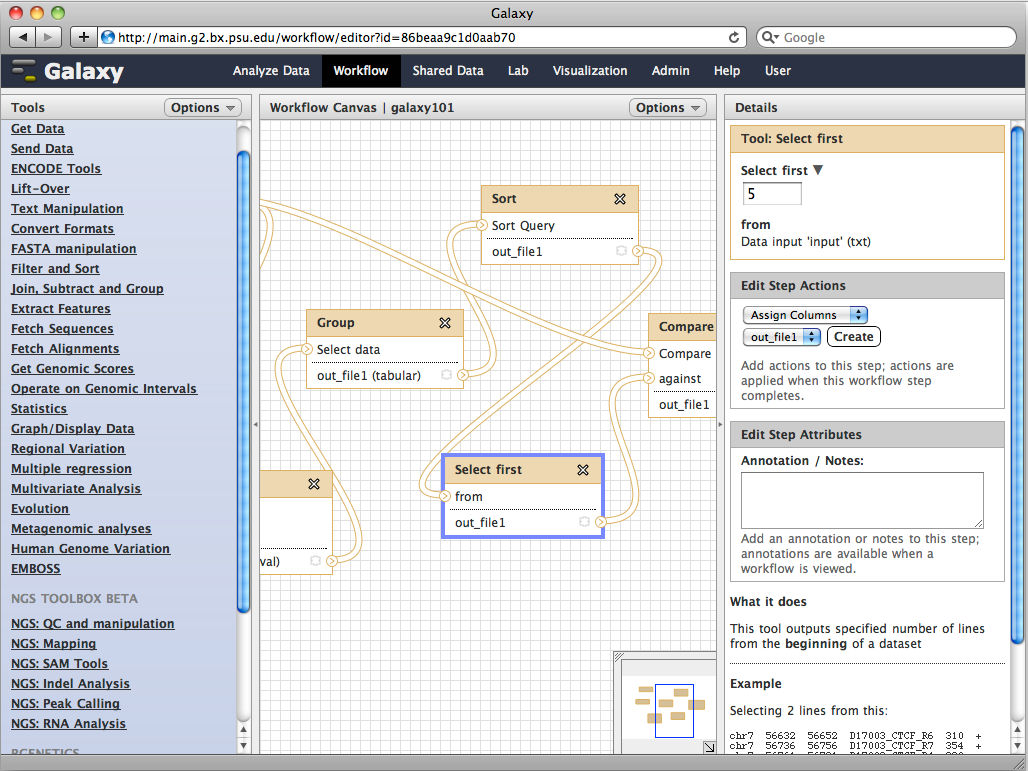
MEME tools contrast with the Galaxy web service [Goecks et al. 2010].
MEME is a more focused tool
chain with a simple point and click interface. Though it can combine tools in a number of useful ways, it has no way to construct
your own pipelines. Galaxy on the other hand is more open-ended, and allows construction of pipelines based on combining existing
tools, and provides a framework for adding new tools.
There are two gaps in current offerings:
- the MEME tools are extremely easy to use, while Galaxy provides greater generality at the cost of more complexity –
the gap here is between MEME’s ease of use and Galaxy’s configurability
- neither web service has a simple model to leverage cloud resources; both can either run on a shared server, with capacity
limits for free use (e.g. the free Galaxy server has a
disk quota per user), or
require installation on the user’s own infrastructure. Galaxy offers expanded use using Amazon EC2,
but requires complex
configuration. To use the MEME tools on cloud infrastructure, you would have to create an instance
yourself.
The aim of this project is to investigate:
- combining the benefits of the MEME and Galaxy services: Galaxy’s configurability and MEME’s ease of use. This requires
finding a new compromise in the design space, with easier to use scripting and pipeline construction. We should test
usability claims versus Galaxy with the help of working biologists; some approaches to investigate include:
- methods for specifying pipelines based on function rather than tools
- methods for documenting workflows that are not purely dumps of scripts and programs run
- methods for specifying tool wrappers by example rather than creating yet another script-like syntax (or yet another
flowchart-like visual workflow notation [Byelas and Swertz 2006])
- transparent transition from local server to cloud-based execution, requiring only payment credentials to use cloud
services.
References
[Bailey and Machanick 2012] Timothy L. Bailey and Philip Machanick, Inferring direct DNA binding from ChIP-seq, Nucleic Acids Research, vol. 40, no. 17 September 2012 pp. e128 (10 pages); first published online: 18 May 2012
[Bailey et al. 2010] Timothy L. Bailey, Mikael Bodén, Tom Whitington and Philip Machanick, The value of position-specific priors in motif discovery using MEME, BMC Bioinformatics, vol. 11 p179, 2010
[Byelas and Swertz 2006] Byelas, H. V., and M. A. Swertz. Visualization of bioinformatics workflows for ease of understanding and design activities, Proc. 6th Int. Joint Conf. on Biomedical Engineering Systems and Technologies, 2006
[Goecks et al. 2010] Goecks, J, Nekrutenko, A, Taylor, J and The Galaxy Team. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences, Genome Biol. 2010 Aug 25;11(8):R86
[Machanick and Bailey 2011] Machanick and Timothy L. Bailey, MEME-ChIP: motif analysis of large DNA datasets, Bioinformatics, vol. 27 no. 12, pp 1696-1697, 2011